A Brief Description
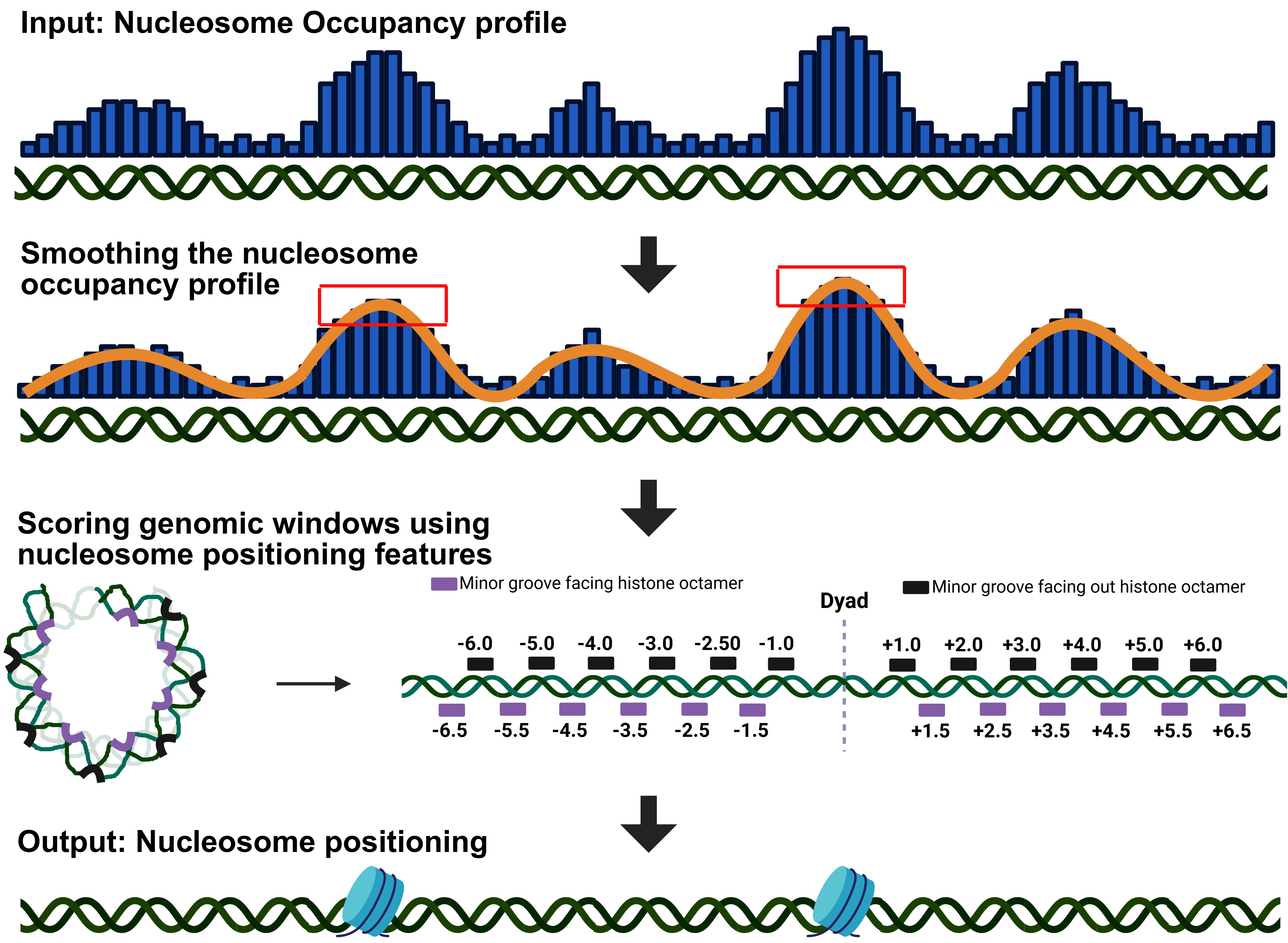
We recently developed a deep learning method that enabled us to identify 88 nucleosome positioning features. This website takes a nucleosome occupancy profile as input and, after smoothing the profile, centers a 20-base window at each local maximum, provided that these maxima do not overlap with other maxima located within 160 bases of each other. Each position within the window is considered a potential dyad position and is scored based on the 88 identified features. The genomic coordinate with the highest score is selected as the dyad position. In addition, the website provides a classifier for nucleosomal and non-nucleosomal DNA and generates a nucleosome positioning score accordingly.
About CCG Lab
We study the associations between key components in the epigenome to understand how its perturbation can lead to cancer. Our team works to identify factors contributing to cancer mutation occurrence in DNA, to discover molecular mechanisms of how mutations and covalent modifications affect nucleosomes and chromatin, their interactions, stability and dynamics.
Go to CCG page